Research Article
Analysis of Transcriptome Sequencing and Related Genes of Flavonoid Biosynthesis from Thesium chinense Turcz 
 Author
Author  Correspondence author
Correspondence author
Medicinal Plant Research, 2021, Vol. 11, No. 4 doi: 10.5376/mpr.2021.11.0004
Received: 22 Dec., 2021 Accepted: 24 Dec., 2021 Published: 31 Dec., 2021
Cao J.F, Zhang Y., Chen P.Y., Wu C., and Deng Y., 2021, Analysis of transcriptome sequencing and related genes of flavonoid biosynthesis from Thesium chinense Turcz, Medicinal Plant Research, 11(4): 1-11 (doi: 10.5376/mpr.2021.11.0004)
In order to find the metabolic pathway of Thesium chinense Turcz flavonoids and related genes, the plants treated with or without silver nitrate (1 mmol/L) were used as test materials, transcriptome sequencing analysis was carried out by using Illumina HiSeq 2000, and analyses of gene functional annotation of unigenes and expression characteristics of key genes for biosynthesis of T. chinense Turcz flavonoids were also performed. The results showed that a total of 69 503 unigenes were obtained by transcriptome sequencing, of which 48 666 were annotated and 9 792 genes were screened by differential gene expression, with an average length of 1 226 bp. Through the metabolic pathway analysis, we found that the expression of CHS, C4H, 4CL, and HCT genes was high, and the expression of PAL, F3H, FLS and FG3 genes was relatively low among the genes closely related to the synthesis of kaempferol and flavonoids. Combined with transcriptome data, the biosynthetic pathway of flavonoid content in T. chinense Turcz was plotted. In addition, 32 008 simple sequence repeat (SSR) loci were determined in the transcriptome of T. chinense Turcz. Dinucleotide microsatellite repeating units were the most abundant in T. chinense Turcz, which accounted for 34.22%. Among the two repeating types of mononucleotide and dinucleotide, the most ascendant repeating units were A/T and AG/CT. This study provided the theoretical basis of molecular pharmacology for improving the yield, the formation of the quality and studying the pathways and related genes of biosynthesis of flavonoids of T. chinense Turcz.
Thesium chinense Turcz is a perennial semi parasitic herb in Santalaceae. The whole grass can be used as medicine. Clinically, it is mainly used to treat pneumonia, pharyngitis, acute mastitis, acute tonsillitis and other diseases. It has strong antibacterial and anti-inflammatory effects and is known as ‘plant antibiotic’ (Zhong et al., 2008). Phytochemical studies showed that kaempferol (also known as thesine III) and kaempferol glycosides and other flavonoids were the main active components in Thesium chinense Turcz. The amount of kaempferol and total flavonoids was often used as an important index to measure the quality of thymus (Lu, 2004). Because of its excellent antibacterial and anti-inflammatory activity, the tablets and capsules prepared from the extract of Thesium chinense Turcz have been widely used in clinical medicine and had great market demand. Thesium chinense Turcz is a semi parasitic plant with small plants, low yield and less resources. Therefore, improving the yield and flavonoid content of Thesium chinense Turcz and looking for new drug sources are the hot spots and urgent problems to be solved. The functional genes related to flavonoid biosynthesis pathway and flavonoid biosynthesis in plants have been widely and deeply studied in a variety of plants (Li et al., 2018; Jiang et al., 2018; Wang et al., 2018; Zhang and Deng et al., 2018; Zou et al., 2019). However, there are no reports on the synthesis mechanism of secondary metabolites of Thesium chinense Turcz. In recent years, the research on Thesium chinense Turcz mainly focused on plant tissue culture (Jiang et al., 2008; Liu et al., 2016; Wu et al., 2016; Luo et al., 2017) and material extraction and separation (Zou et al., 2016; Xuan et al., 2018). Ichihashi et al. (2018) discussed the mechanism of semi parasitism of Thesium chinense Turcz based on transcriptome analysis.
In this study, a high-throughput second-generation sequencing platform was used to construct the transcriptome database of Thesium chinense Turcz. Besides, the transcripts were functionally annotated and the key synthetic genes related to biosynthesis pathway of Thesium chinense Turcz flavonoids were discovered, which laid a foundation for further study on the biosynthesis of flavonoids, cloning and functional verification of key genes, genetic improvement and so on.
1 Results and Analysis
1.1 Sequencing results and assembly
A total of 196 648 transcripts and 69 503 unigenes were obtained from assembly of transcriptome. The average length of transcripts was 1 226 bp, the minimum length was 301 bp, and the maximum length was 17 292 bp. The value of N50 was 2 226, and the value of N90 was 469. The whole transcripts were mainly distributed between 200~6 000 bp, including 44 378 between 300~500 bp (Figure 1).
.png) Figure 1 Statistical analysis of assembly of transcriptome from Thesium chinense Turcz |
1.2 Annotation results of unigenes in different databases
The obtained transcriptome sequences were annotated by database comparison. The number of annotated unigenes was 69 503 (Figure 2), and 35 236 unigenes were annotated in Swiss-Prot (experimentally verified protein sequence database) accounted for 50.69% of the total number of annotations, which was the database with the largest number of annotated unigenes. In the other six databases, NR (NCBI protein sequence database), KOG (eukaryotic lineal homologous gene database), GO (gene ontology database), PFAM (protein family database), NT (nucleic acid sequence database) and KO (gene expression database), the number of unigenes accounted for 46.69%, 27.17%, 49.16%, 49.16%, 39.92% and 23.57% respectively. The number of unigenes successfully annotated in the above seven databases was 7 174, accounting for 10.32% of the total.
.png) Figure 2 Statistics of annotation results of unigenes in different databases |
1.3 Function annotation results of unigenes in 5 different databases
The obtained unigenes were compared with NR, PFAM, GO, NT and KOG databases (Figure 3), and a total of 47 500 unigenes were annotated. Among them, NR database had 68.32% of the total unigenes, and the proportion of unigenes in NT, KOG, PFAM and GO was 58.41%, 39.77%, 71.93% and 71.93% respectively. Unigenes annotated by the above five different databases accounted for 19.35% of the total annotated unigenes.
.png) Figure 3 Statistics of function annotation results in NR, GO, PFAM, NT and KOG databases |
1.4 KOG functional classification of unigenes
After KOG function annotation of unigenes, a total of 18 890 annotation results were obtained. These 18 890 unigenes were classified into 26 functional categories (Figure 4), and these functional categories involved all aspects of physiology and life regulation of Thesium chinense Turcz, of which 2 558 unigenes belong to posttranslational modification, accounting for 13.5% of the total unigenes. There are 2 445 unigenes related to ribosomal structure and biogenesis, accounting for 12.9% of the total unigenes. There are 2 109 unigenes related to general functional genes, accounting for 11.2% of the total unigenes. There are 1 229, 1 445, 1 483 and 1 268 unigenes related to RNA processing and modification, energy production, signal transduction mechanisms and intracellular trafficking respectively, accounting for 6.5%, 7.6%, 7.8% and 6.7% of the total unigenes respectively. Among the total functional categories, there are 27, 84, 124 and 161 unigenes related to cell motility, defense mechanisms, nuclear structure and extracellular structures respectively.
.png) Figure 4 KOG functional classification of unigenes of Thesium chinense Turcz Note: A: RNA processing and modification; B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination and repair; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; O: Posttranslational modification, protein turnover, chaperones; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism; R: General function prediction only; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms; W: Extracellular structures; X: Unnamed protein; Y: Nuclear structure; Z: Cytoskeleton |
1.5 Analysis of GO functional classification of unigenes
After GO function annotation, the unigenes were classified according to three categories: biological process (BP), cellular component (CC) and molecular function (MF) (Figure 5). In the biological process, there were 19 371 unigene annotations related to cellular process, 17 998 related to metabolic process and 14 768 related to single-organism process; In molecular function, there were 18 306 unigene annotations related to binding and 14 739 related to catalytic activity; In cellular component, there were 11 252 unigene annotations related to cell part, 11 252 related to cell, 7 834 related to organelle and 7 649 related to macromolecular complex. The GO function annotation results of Thesium chinense Turcz unigenes can provide guiding information for the further study on the functional genes of Thesium chinense Turcz.
 Figure 5 GO classification of the unigenes of Thesium chinense Turcz Note: a1: Transcription factor activity, protein binding; a2: Metallochaperone activity; a3: Antioxidant activity; a4: Molecular transducer activity; a5: Molecular function regulator; a6: Catalytic activity; a7: Nucleic acid binding transcription factor activity; a8: Transporter activity; a9: Structural molecule activity; a10: Binding, b1: Single-organism process; b2: Growth, b3: Multi-organism process; b4: Detoxification; b5: Cell killing; b6: Reproductive process; b7: Reproduction; b8: Cell aggregation; b9: Developmental process; b10: Behavior; b11: Signaling; b12: Metabolic process; b13: Regulation of biological process; b14: Locomotion; b15: Immune system process; b16: Rhythmic process; b17: Localization; b18: Positive regulation of biological process; b19: Biological phase; b20: Biological regulation; b21: Negative regulation of biological process; b22: Response to stimulus, b23: Biological adhesion; b24: Multicellular organismal process; b25: Cellular process; b26: Cellular component organization or biogenesis; c1: Organelle; c2: Cell part; c3: Nucleoid; c4: Membrane part; c5: Synapse part; c6: Membrane-enclosed lumen; c7: Extracellular region part; c8: Virion part; c9: Extracellular matrix component; c10: Extracellular matrix; c11: Cell; c12: Extracellular region; c13: Virion; c14: Macromolecular complex; c15: Membrane; c16: Synapse; c17: Other organism part; c18: Organelle part; c19: Other organism; c20: Cell junction |
1.6 Analysis of KEGG metabolic pathway of unigenes
KEGG database is a database including system information, genome information and chemical information, which mainly analyzes the metabolic pathway of gene products in cells and the function of gene products. After systematically comparing and analyzing the metabolic pathways of the transcriptome unigene sequence in KEGG database, a total of 16 383 annotation results in 5 categories of metabolic pathways were obtained (Figure 6). Among them, a relatively large proportion was involved in metabolic processes, and there are 8 191 annotations, accounting for 49.99%. It was also shown that there were 5 099 annotations related to genetic information processing, accounting for 31.12%. There were 928 annotations related to cellular process, accounting for 5.66%. There were 348 annotations related to organismal systems, accounting for 2.12%. There were 371 annotations related to environmental information processing, accounting for 2.26%. The analysis results of KEGG database provide a basis for further studying the biological function of the flavonoid metabolism gene of Thesium chinense Turcz and understanding the complex biological behavior of this gene.
 Figure 6 Kyoto encyclopedia of genes and genome annotation classification of the unigenes in the Thesium chinense Turca Note: A: Organismal systems; B: Environmental information processing; C: Genetic information processing; D: Metabolism; E: Cellul arprocesses; 1: Transport and catabolism; 2: Membrane transport; 3: Signal transduction; 4: Folding, sorting and degradation; 5: Replication and repair; 6: Transcription; 7: translation; 8: Amino acid metabolism; 9: Biosynthesis of other secondary metabolites; 10: Carbohydrate metabolism; 11: Energy metabolism; 12: Glycan biosynthesis and metabolism; 13: Lipid metabolism; 14: Metabolism of cofactors and vitamins; 15: Metabolism of other amino acids; 16: Metabolism of terpenoids and polyketides; 17: Nucleotide metabolism; 18: Overview; 19: Environmental adaptation |
1.7 Comparison of unigenes in the NR database
The 69 503 unigenes annotated from Thesium chinense Turcz were compared with the NR database by BLAST (Figure 7). Thesium chinense Turcz has homologous sequences with Quercus suber, Vitis vinifera, Actinidia chinensis, Juglans regia and Nelumbo nucifera, and the homology with Quercus suber and Vitis vinifera was higher, that is 15.2% and 12.1% respectively. The homologous sequences with Actinidia chinensis, Juglans regia, Nelumbo nucifera and other plant species was less than 10%, and the homology was low. The other 65.5% of unigenes belonged to other sequences.
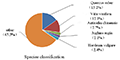 Figure 7 Distribution of species of Thesium chinense Turcz unigenes and NR database BLAST |
1.8 SSR analysis of Thesium chinense Turcz transcriptome
A total of 32 008 simple sequence repeats (SSR) were found in 69 503 unigenes of Thesium chinense Turcz (Table 1). The types of SSR were also rich, and there were 7 436 single gene clusters containing more than 1 SSR locus. Among all SSR loci, di-nucleotide and mono-nucleotide were more, including 16 706 di-nucleotide, accounting for 52.19% of the total, and including 9 690 mono-nucleotide, accounting for 30.27% of the total, which has similar research reports in other plants (Xu et al., 2019). Further analysis showed that there were 156 SSR motif types in the transcriptome of Thesium chinense Turcz. Among the 6 motif types with the highest frequency of repeat motifs, the mono-nucleotide were mainly (A)n and (C)n, that is [A/T (7 593), C/G (2 097)], the di-nucleotide were mainly (AG)n, (AC)n and (AT)n, that is [AG/CT (10 954), AC/GT (3 248) and AT (2 449)], and the tri-nucleotide were mainly (AAG)n, that is [AAG/CTT (989)] (Table 2). The analysis of several motif types showed that with the increase of motif length, its frequency and proportion in the total SSR decreased gradually; The total length of motifs decreased with the increase of motif length. The distribution of SSR in transcriptome sequence of Thesium chinense Turcz showed there were more di-nucleotide (Figure 8), indicating that mono-nucleotide and di-nucleotide were the most active in the whole Thesium chinense Turcz transcriptome. The results of SSR analysis laid a foundation for the subsequent research on the identification, classification and molecular marker development of Thesium chinense Turcz germplasm resources.
.png) Table 1 Statistics of SSR analysis |
.png) Table 2 Distribution of SSR motif type in transcriptome of Thesium chinense Turcz |
 Figure 8 Distribution of SSR repeat motif number of Thesium chinense Turcz |
1.9 Expression analysis of key genes in biosynthesis of Thesium chinense Turcz flavonoids
The main representative components in Thesium chinense Turcz are flavonoids such as thesine Ⅰ, Ⅱ and Ⅲ (Xuan et al., 2018). Among them, thesine III is kaempferol, thesine II is kaempferol-3-O-glucoside, and thesine I is kaempferol-3-O-L-rhamnoside-7-O-glucoside. According to the annotations of the KEGG database, the genes related to the biosynthesis of flavonoids and their corresponding enzymes were screened out, including phenylalanine ammonia lyase (PAL), coumaric acid-CoA enzyme (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), naringenin 3-dioxygenase (F3H), flavonol synthase (FLS), flavonol-3-O-glycosyltransferase (FG3), flavonol-3-O-glucoside L-rhamnosyltransferase (UGT73C6) (Pan et al., 2018). And combining the flavonoids that have been reported in Thesium chinense Turcz, its biosynthetic pathway was speculated (Figure 9). Starting from the synthesis of trans-cinnamic acid by phenylalanine in phenylalanine ammonia lyase (PAL), kaempferol, namely thesine III, was synthesized through a series of enzymatic reactions; Kaempferol synthesized kaempferol-3-O-glucoside (namely thesine Ⅱ) under the action of flavonol glucosyltransferase, and kaempferol-3-O-L-rhamnoside-7-O-glucoside (namely thesine I) was formed under the action of flavonol 3-O-L-rhamnoside-7-O-glucosyltransferase. In addition to kaempferol, Thesium chinense Turcz also contains a very small amount of quercetin. The metabolic pathways of quercetin and kaempferol in plants are very similar. After chalcone forms eriodictyol under the action of chalcone isomerase (CHI), it forms dihydroquercetin through the action of flavonol 3-hydroxylase, and finally forms quercetin under the action of flavonol synthase. Flavonoids widely exist in plants, but the main flavonoids in different plants vary greatly. The expression of key genes in the metabolic pathway of flavonoid biosynthesis is different, and the same key gene also has multiple copies in plants (Liu et al., 2018). Among the key genes of thesine biosynthesis, PAL is the first catalytic enzyme of thesine biosynthesis pathway and the key gene of phenylpropanoid metabolic pathway (Pan et al., 2018). A total of 4 members in PAL family were identified in the transcriptome of Thesium chinense Turcz. Six C4H members belonging to cytochrome P450 (CY450) family were identified. 4CL, which controls phenylpropane metabolism to different metabolic pathways, is the third key enzyme gene, and 14 family members were identified. Two CHS family members were identified, and this enzyme is one of the members of the plant polyketide synthase superfamily and the first key structural gene in the plant flavonoid synthesis pathway (Zou et al., 2019). The subsequent F3H is to convert naringenin into dihydrokaempferol (or eriodictyol into dihydroquercetin) in the plant flavonoid synthesis pathway. Two F3H sequences were identified from Thesium chinense Turcz. Dihydroflavonol is an important substrate for the synthesis of flavonoids and isoflavones. Therefore, F3H is a key enzyme in the synthesis pathway of Thesium chinense Turcz flavonoids, and its product dihydrokaempferol was transformed into kaempferol, namely thesine III, under the action of FLS. FLS is the most important downstream key enzyme in the synthesis pathway of flavonols. Three FLS sequences were identified. FG3 is a key enzyme in the synthesis pathway of thesine kaempferol glycoside, the function of which is to make glycosylate kaempferol (thesine III) transform into thesine II. A flavonol glucosyltransferase gene (FG3, E2.4.1.91) was identified. The number of genes is single, showing high conservation.
 Figure 9 Biosynthesis pathway of Thesium chinense Turcz flavonoid focusing on kaempferol |
1.10 Expression of genes in flavonoid biosynthesis in tissues of Thesium chinense Turcz after different treatments
The biosynthesis pathway of Thesium chinense Turcz flavonoids is directly related to the flavonoid components of secondary metabolites. After the plants of Thesium chinense Turcz were treated with silver nitrate, it was found that the FPKM values of 9 gene enzymes and their corresponding genes with significant differential expression in this metabolic pathway were analyzed (Figure 10). The expression levels of CYP73A (C4H), HCT and 4CL genes were relatively higher, all exceeding 200 (Figure 10). The expression levels of F3H and FLS genes were relatively lower than the previous genes, indicating that the intermediate product of the flavonoid synthesis pathway tends to be only part of the flavonoid synthesis (Liu et al., 2018). PAL is the initial key enzyme in flavonoid biosynthesis pathway, and its gene expression level is relatively low, indicating that PAL is the key rate limiting enzyme in flavonoid biosynthesis. After 6 days’ silver nitrate treatment, the expressions of PAL, C4H, E5.5.1.6 (CHI), F3H and FLS genes were significantly up-regulated. Compared with the untreated group, the expression of enzymes related to flavonoid biosynthesis in the treated samples was higher than that in the untreated samples, The results showed that silver nitrate treatment could significantly affect the expression of enzymes related to flavonoids biosynthesis. In addition, under the action of silver nitrate, the expression of cinnamoyl-CoA reductase (CCR) in plants was significantly higher, and the accumulation of flavonoids in corresponding plants was also increased, which further proved that the expression pattern of CCR gene was related to plant stress resistance (Fan et al., 2015).
 Figure 10 Statistics of expression of key genes in flavonoid biosynthesis Note: CK3: Control check (Third day); CK6: Control check (Sixth day); CK9: Control check (Ninth day); A3: Treated with silver nitrate (1 mmol/L) group (Third day); A6: Treated with silver nitrate (1 mmol/L) group (Sixth day); A9: Treated with silver nitrate (1 mmol/L) group (Ninth day) |
2 Discussion
Thesium chinense Turcz is a perennial parasitic herb. The whole grass can be used as medicine. It shows outstanding clinical efficacy in the treatment of pneumonia, pharyngitis, acute mastitis, acute tonsillitis and other diseases, so its medical value is great. People pay more and more attention to the research and utilization of its medicinal value, the research on its physiology and pharmacology is more and more in-depth, and the research on its related gene is also preliminarily carried out. In this study, the Illumina Hi-Seq 2000 high-throughput sequencing platform was used for sequencing to study the differentially expressed genes in the metabolic pathways of the key active ingredients of Thesium chinense Turcz herb from the molecular direction, and further explore the formation mechanism of the unique active ingredient metabolites of Thesium chinense Turcz herb. A total of 69 503 unigenes were obtained from sequencing, 35 236 unigenes were annotated in Swiss-Prot, 16 383 unigenes were annotated in KO, 32 454 unigenes were annotated in NR, 27 746 unigenes were annotated in NT, 18 890 unigenes were annotated in KOG, which were classified into 26 functional categories, and 34 170 unigenes were annotated in PFAM and GO respectively, Among them, the number of unigenes successfully annotated in the above seven databases was 7 174, and the number of unigenes successfully annotated in at least one of the seven databases was 48 666.
SSR sequence characteristics are important indicators to understand the genomic differences of different species (Shen et al., 2016). In this study, 32 008 SSR loci were identified in the transcriptome sequence of Thesium chinense Turcz, and the highest proportion was di-nucleotide, followed by mono-nucleotide and tri-nucleotide. It was found that the frequency and proportion in the total SSR decreased with the increase of motif length; The total length of motif s decreased with the increase of motif length. The distribution of SSR in transcriptome sequence of Thesium chinense Turcz showed there were more di-nucleotide, indicating that di-nucleotide were the most active in the whole Thesium chinense Turcz transcriptome, which has similar research reports in other plants (Li et al., 2017). The results of SSR analysis laid a foundation for the subsequent research on the development of molecular markers and the construction of genetic linkage map of Thesium chinense Turcz.
The analysis of the research results on the key genes related to flavonoid biosynthesis showed that the expression of genes CHS and CHI was higher than that of PAL in the initial stage of flavonoid synthesis in Thesium chinense Turcz, indicating that PAL gene was one of the main regulation target genes for the regulation of flavonoid metabolism in Thesium chinense Turcz, and the production of flavonoid metabolism can be changed by regulating the expression of PAL gene, which was also verified in Ginkgo biloba (Liu et al., 2018). F3H can catalyze naringenin to form dihydrokaempferol, and then produce kaempferol under the catalysis of FLS. The results showed that F3H and FLS, as the key enzyme genes for flavonoid synthesis, had low expression, indicating that F3H and FLS genes were the key rate limiting genes for restriction of flavonoid products. Therefore, F3H and FLS genes were also the target genes for the regulation of flavonoid metabolism in Thesium chinense Turcz. In addition, the treatment with silver nitrate can improve the expression of the genes realted to flavonoid metabolism and increase the synthesis of flavonoids. The results laid a foundation for the subsequent studies on the regulation of key enzyme genes in kaempferol synthesis and the synthesis of flavonoids by cell engineering of Thesium chinense Turcz. The results of this study will provide a theoretical basis for further research on key gene discovering, metabolic regulation and genetic breeding of Thesium chinense Turcz.
3 Materials and Methods
3.1 Test materials
The Thesium chinense Turcz samples were collected from the experimental base of Dongfeng Town, Wudang District, Guiyang City (106°81’E, 26°64’N), and this medicinal material was identified as Thesium chinense Turcz, a plant of Thesium in Santalaceae, by professor Cao Jianfeng from School of Biological Sciences of Guizhou Education University. One year old Thesium chinense Turcz plants (without branches) naturally grown in the field with 8~12 leaves and basically with the same growth, size and physiological state were selected as experimental plants. They were divided into two groups: normal control group and silver nitrate (1 mmol/L) treatment group. Silver nitrate (1 mmol/L) was sprayed once in the morning and once in the evening, and the samples were taken on the 3rd, 6th and 9th days respectively (the samples in silver nitrate treatment group were marked as A3, A6 and A9 respectively, and the samples in normal control group were marked as CK3, CK6 and CK9 respectively, a total of 6 samples). The samples were stored at -80℃ for later use.
3.2 Transcriptome sequencing
The samples were sent to Novogene Technology Co., Ltd. for transcriptome sequencing and gene function annotation. Oligo(dT) magnetic beads were used to purify mRNA from total RNA, the mRNA was broken into fragments, and a cDNA library of Thesium chinense Turcz transcriptome was constructed. The second-generation sequencer IlluminaHi Seq TM 2000 was used for sequencing after the constructed library passing the quality test, and Trinity software was used to splice De novo transcripts of the obtained sequence. Subsequently, the obtained transcriptome sequence was compared with several commonly used databases through the BLAST program, and the genes related to gene annotation, gene function classification and flavonoid biosynthesis were analyzed (Wang et al., 2018).
3.3 Analysis of key differential genes in the flavonoid metabolism of Thesium chinense Turcz
The annotation information retrieval method was used for database retrieval, and the functional annotation and metabolic pathways of the key genes of Thesium chinense Turcz flavonoids in the KEGG database were analyzed.
3.4 Data analysis
The software MISA (27/09/2010) was used for SSR analysis. The method of SSR analysis referred to http://pgrc.ipk-gatersleben.de/misa/misa.html. After finding out the SSR markers, Primer3 was used for SSR primer design.
Authors’ contributions
CJF was the designer of the experiments and person in charge of the study, guiding experimental design and implementation, data analysis, manuscript writing and revision. ZY participated in experimental design, sample collection, result analysis and manuscript writing; CPY participated in sample collection and result analysis; DY participated in sample collection and result analysis; WC participated in sample collection and result analysis; All authors read and approved the final manuscript.
Acknowledgments
This study was jointly funded by the Natural Science Research Fund Project of Science and Technology Department of Guizhou Province (Qiankehe Foundation [2016]1415); the Natural Science Research Project of Department of Education of Guizhou Province (Qianjiaohe KY [2015] No. 358); the Natural Science Research Fund Project of Science and Technology Department of Guizhou Province (Qiankehe Foundation [2018]1117); First-Class Teacher Team Construction Project of Guizhou Province (Qiangaojiaofa [2017]158) and First-Class Cultivation Science Construction Project of Guizhou Education University (2019YLPYXKB01).
Fan F.F., Li J.Q., Zhan Q.W.,Wang L.H., and Liu Y.L., 2015, Research progress of cinnamoyl-CoA reductase (CCR) gene in plants, Zhonguo Shengwu Gongcheng Zazhi (China Biotechnology), 35(12): 96-102
Ichihashi Y., Kusano M., and Kobayashi M., Suetsugu K.J., Yoshida S., Wakatake T., Kumaishi K., Shibata A., Saito K., and Shirasu K., 2018, Transcriptomic and metabolomic reprogramming from roots to haustoria in the parasitic plant Thesium chinense, Plant Cell Physiol, 59(4): 724-733
https://doi.org/10.1093/pcp/pcx200
PMid:29281058 PMCid:PMC6018956
Jiang Q., Ning Z.Y., Chen J.X., and Liao H.J., 2018, Establishment of somatic cell clones in Thesium chinense Turcz and its in vitro rooting technique, Nongye Kexue Yu Jisu (Agricultural Science & Technology), 9(5): 47-49, 62
Jiang X.H., Yuan J., and Wang X., 2018, De novo transcriptome assembly and annotation of the leaves of Cyclocarya paliurus, Huazhong Shifan Daxue Xuebao (Journal of Central China Normal University (Natural Sciences), 52(6): 822-831
Li L.X., Si S.X., Wei A.Z., Liu Y. L., Feng S.J., and Yang S.X., 2017, Study on development of SSR molecular markers based on transcriptome sequencing and germplasm identification in Zanthoxylum germplasm, Huabei Nongxuebao (Acta Agriculturae Boreali-Sinica), 32(5): 69-77
Li M.X., Wang M., Gan Y.D., Liu Y., Huang Y.J., and Wan C.P., 2018, Transcriptome data assembly and gene function annotation of buds and leaves of Camellia sinensis Cultivar Jing'anbaicha, Xiandai Shipin Keji (Modern Food Science and Technology), 34(5): 93-100, 235
Liu W., Wang J.Y., Li M., Dong J.J., He C.D., Wang G.B., Yu W.W., and Wang Y.Q., 2018, Transcriptome sequencing analysis of gene expression of flavonoid biosynthesis in Ginkgo biloba, Zhongcaoyao (Chinese Traditional and Herbal Drugs), 49(23): 5633-5639
Liu X.Y., Huang Y.F., Wang Z.B., Cao Y.G., and Luo C.M., 2016, Study on the influencing factors of callus culture of Thesium chinense Turcz, Anhui Nongye Kexue (Journal of Anhui Agricultural Sciences), 44(22): 132-134
Lu Y.X., and Wang J.S., 2004, Study on chemical constituents of Thesium chinensis Turcz, Zhongcaoyao (Chinese Traditional and Herbal Drugs), 35(5): 15-17
Luo C.M., Huang Y.F., Liu X.Y., Cao Y.G., and Xie Q., 2017, Optimization of rapid propagation technology on Thesium chinense Turcz, Tianjin Nongye Kexue (Tianjin Agricultural Sciences), 23(11): 11-14
Pan Y., Chen D.X., Song X.H., and Li L.Y., 2018, Transcriptome analysis reveals candidate genes involved in flavonols biosyhthesis in Sophora japonica, Zhongguo Zhongyao Zazhi (China journal Chinese Material Medica), 43(13): 2682-2689
Shen X.B., Yan W.D., Liang X.C., and Sun Z.Q., 2016, Characteristic analysis of microsatellites in Eucommia ulmoides transcriptome, Jingjilin Yanjiu (Nonwood Forest Research), 34(4): 47-51
Wang H., Wang W.W., Wang D.L., Zhang S.H., Hu X.F., Lu S.Y., and Gong X.M., 2018, De novo assembly and functional annotation of transcriptome data of Rhododendron pulchurum cv. Baifeng 4 leaf, Zhejiang Nongye Xuebao (Acta Agriculturae Zhejiangensis), 30(7): 1149-1159
Wu F., Huang Y.F., Cao Y.G., Liu X.Y., Zhu S.Y., and Ren J.X., 2016, The optimization of callus in vitro culture conditions of Thesium chinense Turcz, Tianjin Nongye Kexue (Tianjin Agricultural Sciences), 22(11): 19-23
https://doi.org/10.1007/s12209-016-2565-6
Xu Q.W., Lou X.Z., Yang B., Lin E.P., and Tong Z.K., 2019, Transcriptome sequencing and analysis of Rhododendron fortunei, Zhejiang Nonglin Xuebao (Journal of Zhejiang A&F University), 36(6): 1190-1198
Xuan W.D., Fan Z.P., Hu S.G., and Xu D.D., 2018, Study on hydrophilic chemical constituents from Thesium Chinensis Turcz, Yaoxue Shijian Zazhi (Journal of Pharmaceutical Practice), 36(3): 270-273
Zhong F.L., Wang X.L., Ji P.P., and Zhu B., 2008, Study on extraction process of total flavonoids in Thesium chinense Turcz (Journal of Jilin Institute of Chemical Technology), 25(4): 5-7
Zhang J.E., and Deng H.F., 2018, Analysis of transcriptome sequencing and qRT-PCR analysis of related genes of flavonoid biosynthesis from Eucommia ulmoides calli, Zhongcaoyao (Chinese Traditional and Herbal Drugs), 49(16): 3912-3917
Zou F.X., Xu W., Huang Z.H., and Zhang X., 2019, Analysis of transcriptome sequencing and related genes of flavonoid biosynthesis from Anoectochilus roxburghii, Zhongguo Yaoke Daxue Xuebao (Journal of China Pharmaceutical University), 50(1): 66-74
Zou Y., Hong M., and Yang X.F., 2016, Isolation of chemical components from Thesium chinense, Zhongguo Shiyan Fangjixue Zazhi (Chinese Journal of Experimental Traditional Medical Formulae), 22(7): 74-77
. PDF(651KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Jianfeng Cao
. Ying Zhang
. Peiyu Chen
. Chao Wu
. Yu Deng
Related articles
. Thesium chinense Turcz
. Transcriptome
. Flavonoids
. Biosynthesis
Tools
. Email to a friend
. Post a comment


